Overview
Novel research exploring sustainable plastic degradation garnered high schooler Sanan Khairabadi a finalist position in the 2024 BioGENEius Challenge.
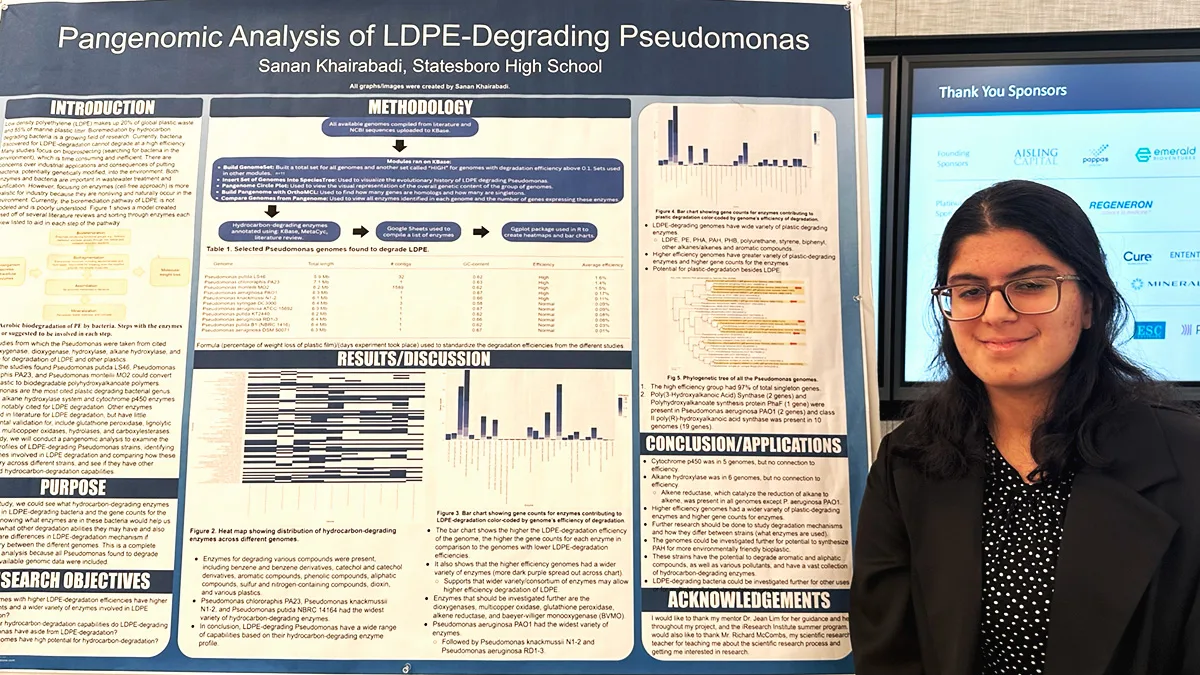
Understanding how bacteria might degrade a form of polyethylene
Novel research exploring sustainable plastic degradation garnered Sanan Khairabadi, a senior from Statesboro High School in Statesboro, GA, a finalist position in the 2024 BioGENEius Challenge. She focused on the enzymes of hydrocarbon-degrading bacteria found to degrade low-density polyethylene (LDPE).
The esteemed competition, founded by the Biotechnology Institute more than two decades ago, showcases the finest young talent in biotechnology from across the United States. Cure hosted the 2024 BioGENEius Challenge ceremonies, awarding a $500 prize to Iyer and three other finalists as well as top prize of $2,500 to Yifan Ding.
The students also toured labs at CURE and met with biopharma executives to discuss careers in the field of biotechnology. Since its inception, the BioGENEius Challenge has seen its alumni go on to impactful careers as entrepreneurs, physicians, healthcare leaders, and champions for sustainable health and environmental solutions.
Khairabadi spoke with Cure about what inspired her, challenges she faced and the journey she took for her project, “Identifying Hydrocarbon-Degrading Enzymes in LDPE-degrading Pseudomonas Through a Pangenomic Analysis.”
This interview was edited for length and clarity.
Cure: Tell us about the inspiration behind your project? What problem were you hoping to solve?
Khairabadi: I was curious to see how the enzymes present for LDPE-degradation differed between [bacterial] strains. Through comparing, I hoped to see interesting patterns in terms of what enzymes are present for LDPE degradation and other hydrocarbon-degrading capabilities across the strains. Through this, we can gain a better understanding of the Pseudomonas that can degrade LDPE specifically.
Cure: What was the biggest challenge you faced while working on your project, and how did you overcome it?
Khairabadi: Since I used KBase, I had to manually search through the list of enzymes for hydrocarbon-degradation. This was challenging because in KBase, most of the enzymes did not have their functions labeled. I had to figure out which enzymes were hydrocarbon-degrading.
To overcome this, I used Metacyc, which showed the pathway for hydrocarbon degradation and the enzymes in each step. [MetaCyc is a curated database of experimentally elucidated metabolic pathways from all domains of life. MetaCyc currently contains 3,153 pathways, 19,020 reactions and 19,372 metabolites.]
I also looked at literature reviews that listed hydrocarbon-degrading enzymes and cited and suggested enzymes for LDPE degradation. From this, I could search for specific enzymes instead of sorting through all the enzymes in the genome, which would take a very long time.
Cure: What was the most surprising or unexpected result you encountered during your research?
Khairabadi: I was surprised that although the [bacterial] strains were in the same genus, some of the strains differed from each other in the enzymes they have for LDPE degradation. In addition, alkane 1-monooxygenase is most cited for degradation, however it was not in all the genomes. For the ones it was present in, there was no difference in how well the genome's degradation efficiency was.
Cure: How do you see the findings from your project being applied in the real world or in future scientific research?
Khairabadi: There is still not an official LDPE degradation pathway. This study showed that although the strains are from the same genus, some of the strains were unique in the enzymes they had for LDPE degradation.
I think future studies could look into how the different strains of Pseudomonas degrade LDPE differently because of their differing degradation enzymes. In addition, the strains with a wider variety of enzymes could be focused on for industrial application and further testing.
Cure: What role did mentorship or collaboration play in the success of your project?
Khairabadi: My mentor introduced me to the topic of hydrocarbon-degrading bacteria and how they aided in the cleanup of the Deepwater horizon oil spill [in the Gulf of Mexico in 2010], and also introduced me to KBase. After learning more about this topic, I did further literature review and became interested in LDPE-degrading bacteria.
Cure: How has this experience influenced your academic or career goals? Do you have specific fields or industries in mind?
Khairabadi: After doing this project, I am aware that there still is a lot more research to do in this field to improve the biodegradation efficiency of LDPE and just in general learning more about the degradation process.
I am very interested in doing work related to sustainability and contributing to sustainability research and efforts after doing this project.
Cure: What advice would you give to other students interested in participating in the BioGENEius Challenge?
Khairabadi: I would encourage them to do a project on something they are passionate about because it makes it enjoyable. Also, if you are interested in pursuing that field of research, doing a project is a great way to further your knowledge.
How Khairabadi describes her project:
My project studies hydrocarbon-degrading bacteria found to degrade low-density polyethylene (LDPE). Several bacteria have been found to degrade plastic. However, they cannot degrade at significant efficiencies.
Most bacteria found to degrade plastic are in the Pseudomonas genus. From my literature review, I found that few studies sequence the genomes of plastic-degrading bacteria, and only three enzymes have been explicitly stated to degrade LDPE. These enzymes include the alkane hydroxylase enzymes, Baeyer-Villiger monooxygenases, and cytochrome p450s.
Most studies do not mention the enzymes contributing to LDPE degradation. They focus on a whole-cell approach rather than a cell-free approach, and focus on bioprospecting for a high efficiency bacteria. There has not been much progress in finding a solution in research and, to my knowledge, I could not find any studies analyzing the genetic variation of hydrocarbon-degrading enzymes in the bacteria.
In my study, I was curious to see the variety of enzymes present in LOPE-degrading Pseudomonas. I downloaded all Pseudomonas with available genomic data on NCBI and uploaded them to KBase, an open software platform developed by the Department of Energy.
From KBase, I ran several modules to visually see the genetic relationships between the Pseudomonas found to degrade LDPE. I identified the enzymes present. My project applies to this track because it helps elucidate what hydrocarbon-degrading enzymes are present in these bacteria that have adapted to break down LDPE, and see whether there are any other hydrocarbon-degrading enzymes present not mentioned in studies but present in other alkane/alkene-degradation pathways.
Follow the BioGENEius Challenge on LinkedIn and our website: Technology Organization Serving in the US | Bio Technology Institute.








